Continous dynamic recrystallization (CDRX)
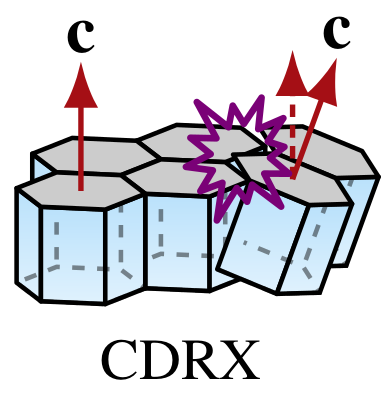
Polygonization (rotation recrystallization, CDRX) accounts for the division of grains along internal sub-grain boundaries resulting from local strain incompatibilities. In effect, CDRX reduces the average grain size upon grain division but does not necessarily change the CPO much (Alley, 1992).
Following Gödert (2003), CDRX can be modeled by approximating this effect as a Laplacian diffusive process on \(S^2\):
where \(\Lambda\) is the CDRX rate-factor magnitude that depends on temperature, stress, strain-rate, etc. (Richards et al., 2021).
Limited use
Notice that this model is currently only relevant to slip-system normals. The model is therefore not yet useful for e.g. olivine.
Matrix model
The corresponding effect on the continuous distribution function is
Code example
import numpy as np
from specfabpy import specfab as sf
# L=8 truncation is sufficient in this case, but larger L allows a very strong fabric to
# develop and minimizes the effect that regularization has on low wavenumber modes (l=2,4)
L = 8
lm, nlm_len = sf.init(L)
### Numerics
Nt = 25 # number of time steps
dt = 0.05 # time-step size
### Initial fabric state
nlm = np.zeros((Nt,nlm_len), dtype=np.complex64) # n state vector
nlm[0,:] = sf.nlm_ideal([0,0,1], 0, L) # sigle maximum at t=0
### Euler integration
for tt in np.arange(1,Nt):
nlm_prev = nlm[tt-1,:] # previous solution
Lambda = 1 # CDRX rate-factor
M = Lambda*sf.M_CDRX(nlm) # CDRX operator
nlm[tt,:] = nlm_prev + dt*np.matmul(M, nlm_prev) # euler step
nlm[tt,:] = sf.apply_bounds(nlm[tt,:]) # apply spectral bounds if needed